Spot the difference
Can we detect when geometric alignment algorithms go crazy?
At NeurIPS 2021, we introduce the first unsupervised, learning based method for detecting changes in image topology. In additioan, we introduce the first dataset of manually annotated changes in topology, which allows us to significantly extend on the evaluation strategies employed in of previous work.
The Problem
Alignment of distributions or images often assumes a common topology across the images, with undefined behaviour if this assumption is violated. Yet in practice, we are often especially interested in these cases where image topology is different or abnormal. For example in medical imaging, the patient we are inspecting is likely in treatment because their anatomy does not follow a ‘standard’, but is abnormal in some way - e.g. by a brain tumor.
Alignment and registration methods that can handle differing topologies exist, but require the prior annotation of the changes in topology - we aim to provide an unsupervised method to detect the changes in topology.
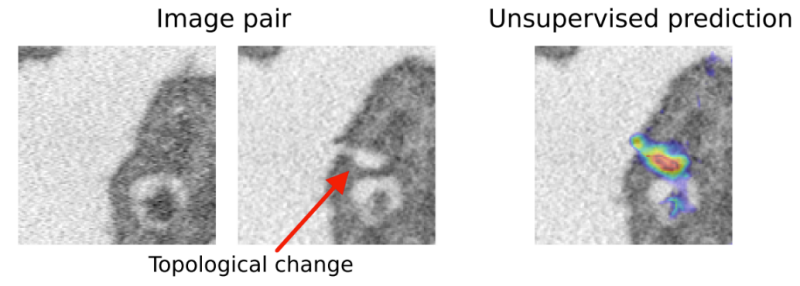
Solution
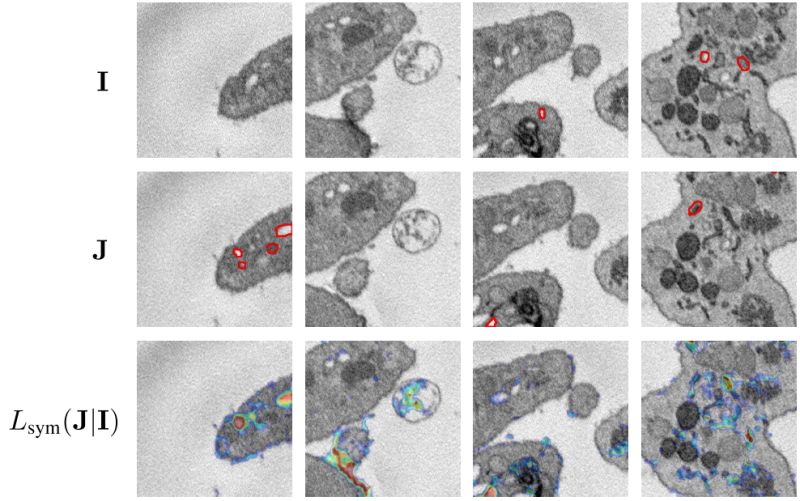
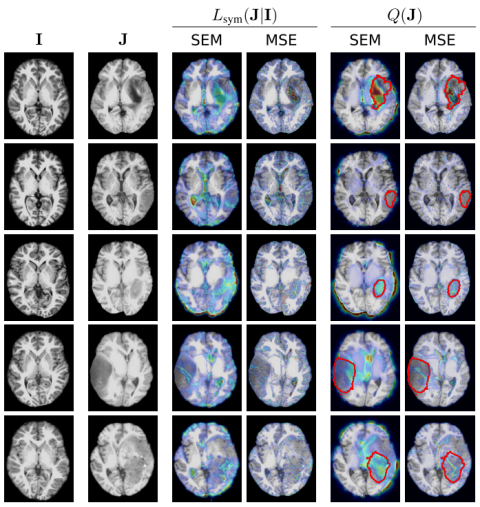
We base our method on probabilistic registration models, using a VAE to give pixel-wise bounds on the likelihood of a diffeomorphic match. We extend prior work by re-defining the output distribution and eliminating hyperparameters in the prior. For validation, we introduce the first dataset with annotated topological changes to aid evaluation in this field, which has been relying on qualitative assessment of small sample sizes and proxy-tasks before.
Results
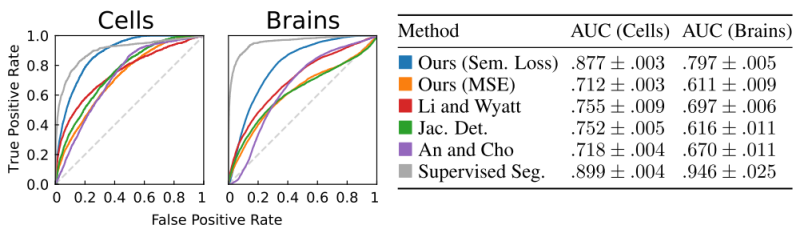
The new method outperforms baselines from topological change detection and anomaly detection by a large margin, and competes with supervised models on one of the datasets.
Code available on Github.